Abstract
Scientific research, especially Neuroscience and Genomics is demanding researchers write code. Many laboratories write their own software to address specific problems no one else has addressed. Empirically, code from papers too many times have not borrowed hones methodologies commonly used by great and productive Software Development teams in corporate America – To improve the code quality and grow the programming skills of their developers. This blog post is focused on Code Review. It is a provides a big picture review of Code Review and get a humble attempt to get researchers interested in it.
Assumptions
This blog post assumes readers work in a wet lab working and write scientific programs to address research questions. It is also assumed that researchers reading are familiar with github. Readers without this background can still understand the key points but may struggle understanding many parts. There are many tutorials on github like TutorialPoint. This blog post is a starting point. Researchers trying to get started with Code Review need to experiment to see what suits their laboratories and do a web search for ideas and available complementary tools.

Introduction
Code Review is a critique of code to ensure quality standards are met. It can be a self-review whereby a researcher examines his/her own code for flaws in quality and areas that can be improved. It can be automated like via linting tool that examines code to ensure proper formatting and structure are used. It can also be done by a human other than the programmer who made the code. Human code review can be done with two or more people studying code (like in Figure 1 above) or offline where a reviewer write notes on a Pull Request.
The benefits of Code Review cannot be overemphasized. It is crucial in growing the skill set of researchers at all levels. Although it adds time to software development timeline, the value-add is tremendous and worth the extra time. It also saves times by likely resulting in less bugs and sharping researchers’ programming skills over time.
I personally read papers in Neuroscience and Genomics. Empirically, Code Review is a step not given due emphasis in the scientific literature. This blog post is an attempt to spark more interest in Code Review and contribute to any discussion of adding it as a a criteria to either accept or reject papers in Scientific Journal. There is no debate against Software Development being an essential part of Neuroscience and genomics research. In turn, there is a crucial need to adopt best practice developed and honed by companies who have built trillion-dollar businesses like Amazon and Microsoft and entire regions like Silicon Valley in California and tech hubs in Austin, TX and Seattle Washington. There are lots of topics but this blog will only address Code Review.
Types of Code review
This blog classifies four types of Code Review – Self (done by the researcher), Automated (done by a linting tool), Peer (done by a person or a group as a prerequisite for considering the code completed and test-ready) and Lab (done as part of a laboratory preparation to submit a paper to a journal.) It is recommended all classes be done. They are listed in order of precedence. Like, it is better to do a self-review before automated. It is better to do automated review before investing a human’s times in a Peer review; and so on. Here are descriptions of the types –
Self Review
It is a grave error to consider code completed and ready to be submitted along with a paper the instance it works. Researchers should take the time to review their own code to determine if it meets quality standards. This save a lot of time and also help reduce bugs as the programmer can catch mistakes and save others from doing that.
Automated Review
There are linters like Pylint that can automate verification of coding standards. Pylint has a set of pre-set rules you can use and you can modify them and/or add new rules. Researchers are expected to satisfy these rules. The rules can be configured to give warning messages so are optional to address or errors which mean they must be addressed. It is best the laboratory get together as a group to determine theses rules based on a consensus. Linting is great in detecting bad code and saves time because it catches issues a human need not worry about. There are many linters for many languages. You can do information-gathering to determine which is best for your needs and how to configure it.
Peer Review
Peer Code Review is done either by two or more researchers meeting and discussing code in person or via a video teleconferencing like on Zoom or Slack or some other system. Or it can offline where a reviewer write comments on code for later reviewer by a researchers. The general idea to to ensure good quality code is met and to provide feedback if any is work needed to obtain this goal. Github has a system where reviewer can comment on a Pull Request. After the review is completed, the reviewer can either request changes on the Pull Request or approve it.
Lab Review
Principal Investigator and lab members review papers before submitting them to journal for publication. It would make sense for Code Review to be part of this process. It would be a good idea for the lab to at least smoke test the code to ensure it does what it is expected to do independent of testing and to ensure the code satisfying the standards of a funding agency like have a specific kind of license or be HIPAA-compliant.
Recommended Code Review by Scientific Journals
Scientific progress has benefited a lot from reviews – Peer review of pre-released papers, reviews of grants, reviews of PhD proposals etc. Researchers at all level typically write proposals and have other researchers, especially more experience researchers and gain valuable feedback. Reviews can provide great ideas the writer has not though of, help clarify ideas, point out fallacies in methodology, help prevent a researcher from wasting years on a poorly designed research project and help increase likelihood of gaining funding. It is recommended you never start an experiment session without a review, especially if you are a PhD student without years of research/real-world experience. It makes sense that code be given this same courtesy.
It would be great if Scientific Journal have a selected linters all submitted code that are part of potentially published papers must satisfy prior to publication. It can be Pylint or a modified version or something else. It makes no difference as long as the linter has strong community support and linting is enforced consistently. Community support is subjective and can be gauged by which potential linters have more stars and forks or some other means like which one has worked well for others. It is automated which reducing the burden on human reviewers.
Like mentioned earlier, it would be a great idea if Peer Reviewers are made responsible also to do a code review. Or a dedicated group of Peer Reviewers do a Code Review on code in papers. This will improve code quality and make code in paper more useful.
Recommended Code Reviews Steps by Laboratories
Step 1 – Self Review
The first test should be a self-review. The researcher can examine at his/her code and check for errors and areas that can be improved. This alleviates the need for others to do this. Linters may take time to run. Therefore, time can be saved if ideally the linter finds no errors or warnings as the researcher can progress to the next step. Otherwise, the code has to be improved and the linter re-ran.
Step 2 – Automated review
Linter are automated tools that check code for quality based on configured rules. There are a variety of linters for a variety of popular programming languages. The beauty of linters is that they automate code review, so free human code reviewers to focus less of styling and more of logarithms and efficiency. You can do information-gathering on the web on how to set one up one for your particular code.
To illustrate the core idea of linters with a simple example, imagine this is code a researcher needs linting performed on. Typically it is best to have lint as part of Continuous Integration. Smaller code may have this as part of an IDE. In a JupyterLab/Jupyter Notebook application, you can try this approach. To keep things simple, let’s try this code snippet from Code Snippet 1 –
def apple_engine():
return ['Apple', 'orange', 'pear'];
Code Snippet 1. Code that fails linting.
The code in Code Snippet 1 fails the linting. It is bad practice to use semi-colon in Python in general. The researcher needs to remove the semi-colon at the end of the line. So the researcher does code in Code Snippet 2-
def apple_engine():
return ['Apple', 'orange', 'pear']
Code Snippet 2. Code that passes linting.
Now, linting passes and the code is ready for a human to review.
Step 3 – Performing Manual Code Review
There is no hard-and-fast rules. Empirically, I leave styling up to linter. Code Review by humans examine the logic and flow of the code. Comments are made on anything questionable. There are two grades – Approve or Request changes. Approve means any change recommendations are optional and the code looks fine. Request changes means the researcher is required to make at least one change. The focus is typically on a single Pull Request and not the code base overall structure like Readme.md files and license.md file.
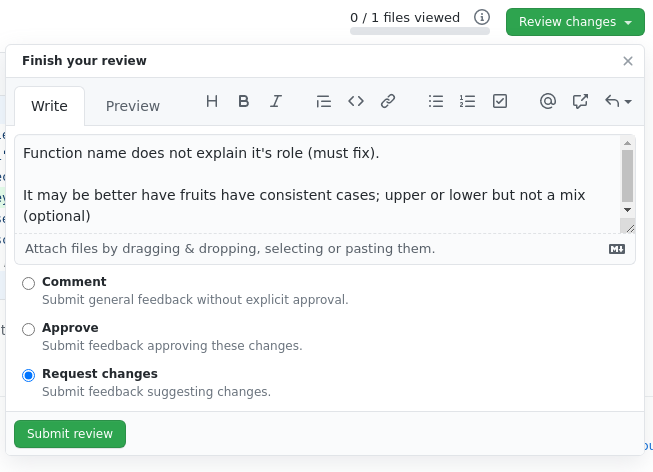
The reviewer doing the Code Review will see Code Snippet 2 in a Pull Request. The code is not of good quality because the function’s name is not intuitive. This is a change the researcher must correct in order to pass the review. The code reviewer can also warn the researcher that fruit names should be in a consistent case, but marks this as an optional change. If this is done offline, the reviewer can comment on the Pull Request as illustrate here to convey this to the researcher. The “Finish your review” modal popup can say something like –
The researcher makes changes. The function name is now more descriptive. The casing was untouched but does not matter as changing it was optional. The code change is illustrate in Code Snippet 3 below –
def get_list_of_rat_feed():
return ['Apple', 'orange', 'pear']
Code Snippet 2. Code that passes linting.
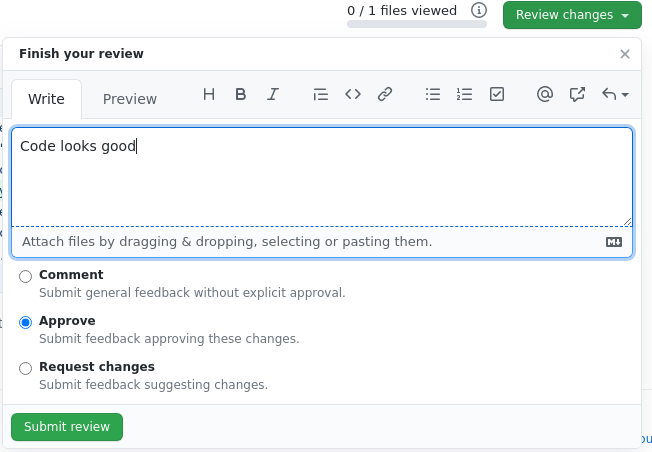
The reviewer satisfied with the change can approve the code as illustrated in Figure 3.
Code Review takes times, but the benefits more than makes up for it. The researcher learned not to use semi-colons to end a line in Python and name functions appropriately. Teaching is a great way to reinforce concepts and the reviewer benefits. The teams benefits as code quality was improved. This example does not illustrate Code Review catching bugs. But empirically, the team I worked on that produced the least bugs is the team that has mandatory Code Review. This has saved time and enhances user experience.
This is a trivial example but illustrate the core ideas of Code Review and a workflow. Some code reviewers smoke test code and read the task to better understand the requirements and ensure the code satisfies them. But this is optional and testing can be left to the testing team. It is ideal not to use the code reviewer as tester, although in small teams there may be no choice.
Step 4 – Lab review
Lab Review focus more on the overall code base like Readme.md, license.md, ensuring there is a mean for other researchers to access the data and other factors that are needed to ensure grant requirements are satisfied. This is primary done by the Principal Investigator or some responsible for compliance like a director. It tend not to be too focused on Pull Requests. This also has no hard-and-fast rules and varies per lab and per grant.
Personal Anecdote
I foresee a future where big research breakthroughs are made possible by code. I truly believe in this. It is why I left Chicago to move to Silicon Valley in California – To be around smart people who push the envelops of programming, Neuroscience and genomics. Silicon Valley has among the smartest people on the planet and it is great to be near them.
I am proud to have contributed to the growth of others by doing Code Review for them. It is time well spent and I will never work in a laboratory where Code Review is not standard. When I do code reviews, I am brutally honest and demand the next review to go better if one is needed. I expect others to do the same for me. It is important to be honest and have frank and open communication to helps people grow. A culture of gossiping about others behind their back while not providing them feedback does not help growth, is counterproductive and a waste of time. I recommend leaving such as job rather than try to salvage an upcoming disaster the company/group may become. I know first hand why not to make that mistake. Sometimes programmer do approaches the reviewer has no experience in so it is good to talk to the programmer to discuss why an approach was taken rather than conclude the programmer does not know what he or she is doing and make other believe so too. Code Review is an opportunity for both the programmer and the reviewer to learn. With honest Code Review, egos can get bruised the benefits of learning and growth heals the wounds. I bring this up because the beauty of Code Review will be lost if open and honest communication is hindered by political correctness.

Additional Topics
There are many other topics to consider in improving the quality of code in scientific papers. A few are –
- Documentation-comments via docstring or an equivalent. This helps in creating automated documentation like Sphinx
- Having a thorough Readme that helps other researchers understand how to use the code
- Try to contribute code as a plugin to a larger system. If approved, it will encourage others to reuse the code and make modifications. There is too much scientific code written and used just once.
- Setting up code to encourage others to contribute, like making a contributing.md file
- Run linting with a Continuous Integration system like Travis CI or Circle<a href=”http://<!– wp:list –> <ul><li>Documentation-comments via <a href=”https://www.programiz.com/python-programming/docstrings#:~:text=Python%20docstrings%20As%20mentioned%20above%2C%20Python%20docstrings%20are,can%20access%20these%20docstrings%20using%20the%20__doc__%20attribute.”>docstring</a> or an equivalent. This helps in creatinig documentation</li><li>Having a thorough <a href=”https://docs.github.com/en/github/creating-cloning-and-archiving-repositories/creating-a-repository-on-github/about-readmeshttps://docs.github.com/en/github/creating-cloning-and-archiving-repositories/creating-a-repository-on-github/about-readmes”>Readme</a> that helps other researchers use code </li><li>Try to contribute code as a plugin in a larger system. If approved, it will encourage others to reuse the code and make modifications. There is too much scientific code written and use once.</li><li>Setting up code to encourage others to <a href=”https://docs.github.com/en/communities/setting-up-your-project-for-healthy-contributions”>contribute</a>, like make a <a href=”https://mozillascience.github.io/working-open-workshop/contributing/”>contributing.md</a> file</li><li>Run linting with a <a href=”https://en.wikipedia.org/wiki/Continuous_integration”>Continuous Integration</a> system like <a href=”https://www.travis-ci.com”>Travis CI</a> or Circle CIAutomated test (Unit, Integration, BDD, etc)</li></ul> CI. They are complementary for small project. Your university or Research institute may have a licence which you can use.
- Have Automated test (Unit, Integration, BDD, etc)
Conclusion
Code Review is a great way to enhance research quality and better help researcher learn good programming habits. The are four categories of Code Review that involve automated or manual verification. The time it takes to do Code Review is worth it as there can be tremendous gains in code quality and improving the programming skills of researchers. It would be great if Scientific Journals require linting of all code that is part of a papers.
References/Further Reading
- Code Review Wikipedia page. Access July 25th, 2021. https://en.wikipedia.org/wiki/Code_review
- Lint (Software) Wikipedia page. Access July 25th, 2021. https://en.wikipedia.org/wiki/Lint_(software)
- Writing comments on a Pull Request. https://docs.github.com/en/github/collaborating-with-pull-requests/reviewing-changes-in-pull-requests/commenting-on-a-pull-request
- TutorialPoint github tutorial. https://www.tutorialspoint.com/git/git_basic_concepts.htm#:~:text=%20Git%20-%20Basic%20Concepts%20%201%20Version,is%20available%20freely%20over%20the%20internet.%20More%20
- Pylint. Accessed July 25th, 2021. https://pylint.org
- Continuous Ingeration Wikipedia page. Accessed July 25th, 2021. https://en.wikipedia.org/wiki/Continuous_integration
- Stackoverflow answer in getting Pylint in Jupyter Notebook. Accessed July 25th, 2021. https://stackoverflow.com/a/54278757/178550